Research
Higher order Cellular Functions
Membrane proteins including receptors, the cytoskeleton, and intracellular vesicle transport play a pivotal role in maintaining cell morphology, polarity, adhesion, and differentiation under the control of a complex signal transduction network. In order to understand higher order cellular functions, we have to know functions of signaling proteins and how these proteins are regulated in a whole cell system. We tackle these problems using plants, model organisms, and animal models, from the viewpoint of molecular genetics, cell biology, physiology and biochemistry.
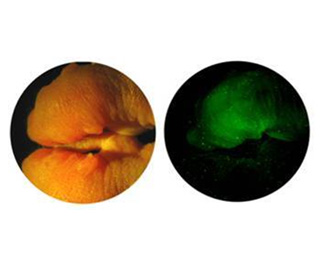
Plants are commonly considered as inanimate and “still life” objects, however, we are simply limited in our abilities to directly sense their movement in our temporal sense. Upon close evaluation with time-lapsed photography, it becomes evident that plants are very animated and their dynamic movement and development is revealed. The plant hormone “Auxin” has a central functional role in plant morphogenesis. The main focus of this laboratory pertains to the study of “Early-Auxin Inducible Genes” and to investigate the functional roles of auxin in the synthesis of new organs (Organogenesis). We primarily utilize Arabidopsis as a functional model system for both forward and reverse genetics. In addition, we also use both potato and carrot as additional models.
 |
Masaaki K. Watahiki (Associate Professor) |
| Keywords | Auxin, feedback regulation, gene expression, molecular biology, plant physiology, genetics, plants, Arabidopsis, carrot, potato |
Plants possess multiple attractive features and potential that we, as humans, simply lack. For example, plants are readily capable to regenerate an entire organism from a single cell by using only light, water and micronutrients. In addition, stem cells for most plant species possess capabilities for indefinite cell proliferation. Plants are capable of surviving exposure to dynamic seasonal conditions from summer to winter. While plants are actively photosynthesizing, plants will never die from “cancer-like” effects even in continuous exposure to sunlight. Over the course of millions of years, plants have evolved many unique and beneficial characteristics which have the potential to bring great value for human beings and our mother earth. One of our primary goals is to unveil these powerful attributes of plants or plant cells and to learn their underlying mechanisms. As a result, these findings may enable us to develop novel plants which are capable of enhanced growth and productivity even under harsh environmental conditions.
 |
Tomomichi Fujita (Professor) |  |
Prerna Singh (Assistant Professor) |
| Keywords | Molecular & cell biology, Bryophytes, cell proliferation and differentiation, environmental response, phytohormone, cell polarity, asymmetric cell division, cell cycle regulation, cell-cell communication, perception and mind in plant life, space utilization science, space moss, auxin polar transport, imaging, body axis formation, Cell wall, Osmotic pressure, Biomechanics, Micropillar, Microfluidics,terraforming |

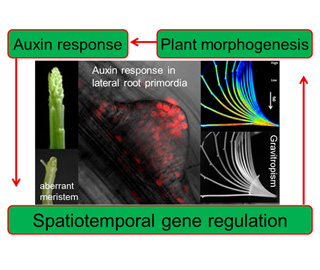
Plants acquire highly diverse morphologies during evolution. This diversity is a consequence of the plant's unique principle of growth and development, which is governed by a genetic program, but is modulated by environmental signals. Polar auxin transport, which is controlled by the polar localization of the auxin efflux carrier PIN protein, acts as the basis for this principle. The polar localization of PIN is regulated by various cellular factors, including membrane trafficking, membrane fluidity and cytoskeleton. Moreover, PIN localization is also regulated by environmental signals, including light and gravity signals, as well as the starvation and salinity stress. Because of this complexity, the mechanisms that regulates the polar localization of PIN remain elusive. We introduce cellular and molecular biological techniques to elucidate the mechanisms of polar localization of PIN. Employing mathematical and theoretical biological techniques, we elucidate the relationship between the evolution of the molecular basis of PIN localization and the morphological diversification during plant evolution. In our experiments, variety of plants, including seed plants, ferns, mosses and algae will be used.
 |
Satoshi Naramoto (Associate Professor) |
| Keywords | morphogenesis, polar auxin transport, body axis, cell polarity, membrane domain, cytoskeleton, membrane trafficking, environmental response, stress evolution, diversity, light response, gravity response, live cell imaging |

Environmental Life Science
All living things are exposed to environmental stresses. Understanding the physiological processes that underlie stress injury and the mechanisms by which living organisms adapt and acclimatize to environmental stress is important to achieve a good understanding of basic concepts in biology. We will study, through lectures, how living organisms sense and respond to environmental changes, which finally lead to reconstitution of the organism. Subjects covered in the course will include: 1) energy conversion systems for photosynthesis and mass production, 2) metabolic regulation controlled by functional RNA species and post-translational processes, such as ubiquitin-mediated protein degradation, and 3) cellular regulation, such as cell division, cell differentiation, and development triggered by chromatin remodeling, gene regulation, and growth regulators.
For many years, DNA was known as the key cellular component which captured encoded genetic information and served as the template for active cellular components; beginning with RNA and subsequently functional proteins. However, active RNA molecules and epigenetic modifications have been recently shown to play critical and highly specific regulatory roles for important biological activities. By using plants as our model system, we study the functional mechanisms of non-coding RNAs and epigenetic regulation. We are very interested to understand how the effects of the epigenetic regulation impact the capability of plants to adapt to their environment. A main objective of our studies is to identify novel gene regulation factors. Secondly, we also aim to develop enhanced plants through application of the identified regulation factors.
 |
Hidetaka Ito (Associate Professor) |
| Keywords | environmental adaptation, transposon, plant molecular genetics, epigenetics, plant breeding, environmental stress, evolution |

Plant growth and development are controlled by the concerted actions of many
signaling pathways which are triggered by developmental and metabolic cues.
Nutrients such as sugars and nitrogen compounds play an important role as
signaling molecules that modulate the expression of many plant genes involved in
diverse physiological processes, such as germination, seedling development,
flowering, senescence, and pathogen responses. Our goal aims to clarify
signaling networks which mediate nutrient utilization and plant growth regulation.
 |
Takeo Sato (Associate Professor) |
 |
Junpei Takagi (Assistant Professor) |
| Keywords | environmental adaptation, molecular biology, plant science, ubiquitin- proteasome system, metabolic regulation, plant immunity, proteome, biotic and abiotic stress, organelle function, live cell imaging |
The control of gene expression is exerted by multiple steps such as transcription, RNA
processing and export, mRNA stability, translation and post-translational events. The recent discovery of biologically active small RNAs has enhanced the impact of post-transcriptional regulation, and in particular, the alterations of mRNA stability on gene regulation. The control of mRNA turnover is thought to be an important component of the rapid response to environmental changes. The goal of our projects is to demonstrate the importance of mRNA turnover control in regulating gene expression in response to several stress conditions in plants.
 |
Yukako Chiba (Professor) |
| Keywords | stress response, regulation of gene expression, mRNA degradation, Arabidopsis, molecular biology |
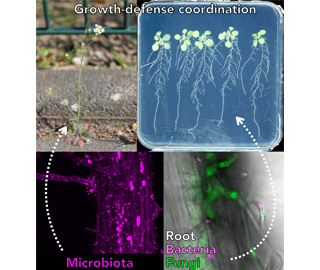
Plants in nature are exposed to a massive variety of microbes. Just like human health relies on the appropriate coordination of gut microflora, plant physiology is immensely influenced by its associated microbial community (the plant microbiota). Our group focuses on the molecular dialog between plants and microbiota and aims to understand the molecular mechanisms by which plant microbiota interfere with plant development and immunity. Our ultimate goal is to describe the true art of life of plants in nature at the molecular level by exploring plant-microbe interaction and its evolution.
 |
Ryohei Thomas Nakano (Professor) |  |
Tomohisa Shimasaki (Assistant Professor) |
| Keywords | Plants, microorganisms, plant microbiota, plant immunity, plant development, inter/intra-cellular signaling, plant-microbe interactions, plant pathology, biochemistry, molecular genetics, cell biology, molecular ecology, bioinformatics |
Animal Behavior and System Control
Functions of the nervous system, including sensory integration, motor control, learning, memory, and motivation, are closely associated with animal behavior at the level of the organism. Analyses of the neuronal and circuit operations underlying these functions are conducted at the level of integration by directly relating them to organism-level behavior. Students are expected to develop a systematic understanding of the leading-edge knowledge and concepts attained by state-of-the-art experimental techniques, including molecular biology, biophysics, neuroendocrinology, and systems physiology, as well as techniques for reconstruction of nervous functions by computer simulation together with its significance in biological research.
When animals receive sensory stimuli from environment, they perform appropriate behaviors via sensory perception, decision making, and motor planning that are mediated by their central nervous system. By using a combination of behavioral analyses, electrophysiology, optical imaging and machine-learning approaches, we address understanding the neural basis underlying wind-elicited behavior in the cricket as a behavioral model. The ultimate goal of our research is a complete understanding of whole neural circuitry and computational process from sensory perception to motor output in specific behaviors. In addition, we started new research on electric fish, which have specialized electric organ and electrosensory organs. We study the neural mechanisms underlying electric communication, including sensory processing and motor control, and the evolution of electrosensory systems.
 |
Hiroto Ogawa (Professor) |  |
Matasaburo Fukutomi (Assistant Professor) |
| Keywords | insect, behavior, brain, nervous system, neural circuit, electrophysiology, imaging, neuroscience, neuroethology |

Our research incorporates both mechanistic and functional approaches to address the question why living things show such a great diversity in behaviors and their underlying cognitive functions. We focus primarily on functional aspects and we investigate the behavioral ecology of passerine birds (mainly Estrildid finches) with particular interest in social communication; including courtships and parent-offspring interactions,
reproduction, development and life history.
 |
Masayo Soma (Professor) |
| Keywords | songbird, social behavior, development, learning, courtship, cognition, ethology, behavioral ecology |

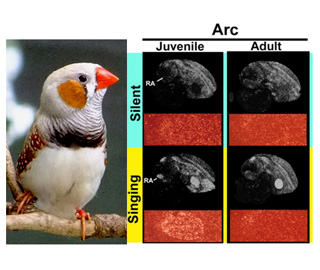
Animal behavior is influenced by both environmental and genetic factors. However, much remains to be learned regarding how and when the environmental and genetic
factors act and how developing behavior itself affects the molecular basis in the neuronal substitute. The primary focus of our laboratory is to elucidate these questions by using songbirds as an animal model for vocal learning and to identify its critical period.
 |
Kazuhiro Wada (Professor) |  |
Noriyuki Toji (Assistant Professor) |
| Keywords | animal behavior, gene expression, learning and memory, individual difference, songbird, communication, epigenetics |
Our goal is to reveal (i) how neuronal circuits work as a system to process sensory inputs, (ii) how this processing is modulated by a specific environment or by the mental state or the condition of the body, (iii) how behavioral decisions are made according to previous experiences and the current situation, and (iv) how these processes depend on the dopaminergic and octopaminergic systems. The answers to these questions will reveal how an animal displays adaptive behavior in response to a given situation that changes every moment or even develops intelligent behavior to cope with a difficult situation, and also help us invent biologically inspired sensors and environmental-friendly pest control. To study these problems in our laboratories, we are utilizing invertebrate animals such as insects and cephalopods.
 |
Nobuaki Tanaka (Associate professor) |
 |
Hiroshi Nishino (Assistant Professor) |
 |
Michael Schleyer(Assistant Professor) |
| Keywords | Drosophila melanogaster, cephalopod, pygmy squid, sensory processing, neurophysiology, neuroanatomy, genetics, behavioral studies, pest control, biomimetics |

Humans use only one hand for performing most tasks in various situations. Such lateral preference in behaviors has been widely documented in many vertebrates and invertebrates. The dominant side may have possibly better locomotion performance (accuracy and/or speed) than the non-dominant side. However, the differences in the nervous systems between righty and lefty, the developmental process of dominance, what genes and molecules regulate dominance, and when it was established evolutionarily still remain essential mysteries. Using African scale-eating cichlids, which are known for their pronounced handedness, I am investigating the mechanisms governing laterality from multiple approaches.
 |
Yuichi Takeuchi (Associate Professor) |
| Keywords | Laterality, Intraspecific polymorphism, Learning, Phenotypic plasticity, Neural circuits, Development of behavior, Evolution, Gene expression |

If we sleep eight hours a day, we spend as much as one-third of our lives sleeping. Why do we sleep? Why do we dream? Why do we have rapid eye movement (REM) and non-REM sleep? We are still unable to correctly answer such fundamental questions. In our laboratory, we perform research to answer these questions using genetically engineered mice. We aim to clarify the neural mechanisms and physiological functions of dreams, in particular.
 |
Tomomi Tsunematsu (Associate Professor) |
| Keywords | Sleep, Dreams, Neuron, Memory, Genetically engineered mice, Electrophysiology, Optical imaging, Optogenetics, Programming |
Reproductive and Developmental Science
An important general goal of life science research is to elucidate the factors regulating the formation of germ cells and how a new individual is generated after fertilization. This knowledge can be applied directly to various reproductive manipulations with direct consequences for our life, such as in vitro fertilization, contraception, and production of useful crops. Reproductive and developmental biology have two aspects: pure science, which pursues the mechanisms guaranteeing the continuity and diversity of life; and applied science, which develops technology to artificially control reproduction and development. Social interests in this academic field, as represented by cloned animals and regenerative medicine, are now very high.
In this field, we are studying the general mechanisms of the formation and maturation of germ cells and regulation of cell division and differentiation in development.
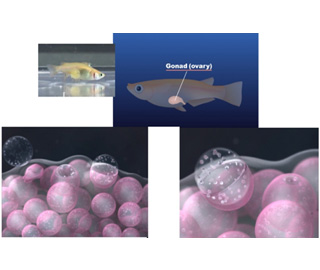
The supply of germ cells (oocytes) to produce the next generation is the most important function of the ovary. We are studying the unsolved issues associated with ovarian functions. We are analyzing to elucidate the mechanism of oogenesis, ovulation, and tissue repair after ovulation using mouse, medaka, and zebrafish.
 |
Katsueki Ogiwara (Associate Professor) |
| Keywords | follicle selection, ovulation, tissue repair |
Steroid hormones regulate physiological responses in vertebrates by binding to the nuclear receptors (NR), a ligand-activated transcription factor. We study the evolution of vertebrate NRs and investigate how steroid hormones act in many vertebrates. We also analyze the temperature-dependent sex-determination/ differentiation of reptiles, and the effects of endocrine disrupting chemicals; including the evaluations of pharmaceuticals on animals.
 |
Yoshinao Katsu (Professor) |
| Keywords | endocrinology, steroid hormone, nuclear receptor, endocrine disruptor, sex determination/differentiation, |

How is sex determined in animals? We are interested to study sex determination, regulation of chromatin, fertilization in both mammals and birds. Using the extremely rare genus Tokudaia, which is mammals but lacks a Y chromosome, we aim to elucidate new sex determination genetic mechanisms and understand the evolutionary significance of the loss of the Y chromosome. We also aim to discover mechanisms unique to birds in ovary and testis determination using birds such as chickens and emus.
 |
Asato Kuroiwa (Professor) |  |
Ikuya Yoshida (Assistant Professor) |
| Keywords | Keywords: sex determination, heterochromatin, EC cell, gene, Y chromosome, bird, mammal, reproductive developmental biology, molecular biology, |

Laboratory of Reproductive and Developmental Biology - (Mizushima Lab)
The life of an organism begins at fertilization, and in the case of birds, primordial germ cells (PGCs), which are the precursor of germ cells, are formed just after fertilization. Fertilization in birds exhibits polyspermy, in which multiple sperm successively penetrate the egg at different points, whereas fusion with the female nucleus involves only one selected sperm nucleus. However, it has been shown that the ooplasm signaling generated by other supernumerary sperm in the egg plays a role in initial formation of PGC. Our research group is conducting studies to elucidate the intracellular signaling. The effects of endocrine disrupting chemicals on early PGCs were also analyzed.
 |
Shusei Mizushima(Associate Professor) |
| Keywords | birds, early development, fertilization, molecular biology, primordial germ cell |

Most of genome sequences are nongenic regions that regulate gene expression and are transcribed as noncoding RNAs. We aim to elucidate the physiological significance and molecular mechanism of genome sequences in reproduction. Our main projects are multifunctional genome elements in testis, long noncoding RNAs in testis and ovary, and the evolution of regulation of oogenesis.
 |
Atsushi P. Kimura (Professor) |  |
Chika Fujimori (Assistant Professor) |
| Keywords | molecular biology, reproductive biology, endocrinology, evolution, gene regulation, epigenetics, long noncoding RNA, multifunctional genome, spermatogenesis, ovary |
A multicellular organism begins with a single cell, the fertilized egg, and subsequently undergoes differentiation and morphogenesis to become an embryo. All cellular functions and patterning decisions that occur prior to the activation of the zygotic genome depend on maternal factors deposited in the egg during oogenesis, which become active after fertilization. By using zebrafish and mouse as model systems, we investigate the deposition of maternal factors in oocytes that promote developmental processes. Our aim is to identify novel mechanisms of oogenesis and developmental processes by isolating and analyzing maternal factors.
 |
Tomoya Kotani (Associate Professor) |
| Keywords | vertebrate, oocyte, egg, early development, maternal factor, cell biology, molecular biology, molecular genetic |
Laboratory of Animal Genetics
Completion of the genome sequence project and development of next-
generation sequencing technique have dramatically changed a design of
research strategy in biomedical science. However, functions of all genes in the
whole genome are not well understood even now. We elucidate functions of
disease-related genes by use of gene-manipulated rats and mice generated in
our laboratory.
 |
Kazuhiro Kitada(Associate Professor) |
| Keywords | Animal models for human diseases, gene manipulation, rat, mouse |